A framework for characterizing dendritic cells
Immunological sentinels known as dendritic cells (DCs) help the body eliminate a wide variety of potential threats, from pathogens to cancer—but they are not all created equal. Some DCs are better at fighting bacteria, others at combating viruses, and still others for keeping tumors at bay. Unfortunately, the existing framework for classifying DCs is confusing and inexact, which makes cross-disciplinary research difficult.
A technique developed at A*STAR will help to distinguish the various DCs and ensure that everyone working on basic and applied immunology is talking about the same things. This will ultimately facilitate future immunotherapies against cancer and infectious disease.
"The big limitation of the field for the past 10 years has been to define these subsets of dendritic cells in different tissues and across species," says Florent Ginhoux from the A*STAR Singapore Immunology Network, who co-led the research. "This study aimed to categorize these subpopulations of cells so that all of us, from basic immunologists like me to clinicians engaged in translational research, have a common language."
All DCs arise from stem cells in the bone marrow, but they can go one of two ways. They can become conventional DCs (cDCs) that patrol tissues for foreign invaders, chew up the intruders, and present bits of the enemy for immune T cells to recognize and destroy. Or they can become plasmacytoid DCs (pDCs), which spew out large amounts of immune-activating interferon proteins to further stimulate T-cell attack in the face of viruses.
Three years ago, Ginhoux and an international group of immunologists proposed a new nomenclature system in which cDCs were further divided into two lineages, one that mounts the type of immune response seen against intracellular pathogens as well as tumors, and the other that reacts to extracellular pathogens such as fungi and bacteria. It was a provocative idea—and one that Ginhoux's team needed to prove experimentally.
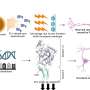
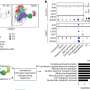
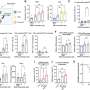
So the A*STAR researchers, in collaboration with others in Europe and elsewhere in Singapore, used sophisticated cell sorting techniques on multiple tissues from mice, monkeys and humans to suggest a panel of surface proteins that clearly distinguished the different DC types. These protein markers also helped demarcate macrophages, a related type of immune cell that often gets mixed up with DCs.
Now that the groundwork for identifying DCs is laid, the real applied research can begin. "Using this blueprint," says Ginhoux, "we can study samples from different disease states to see if there are signatures of dendritic cells before and after treatment."
More information: Martin Guilliams et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species, Immunity (2016). DOI: 10.1016/j.immuni.2016.08.015