Using genetics of human fat cells to predict response to anti-diabetes drugs
Drugs called thiazolidinediones (TZDs) reverse insulin resistance in patients with type 2 diabetes by targeting the activity of a receptor protein. However, an array of side effects, including weight gain, edema, and high cholesterol, limits their use in the clinic.
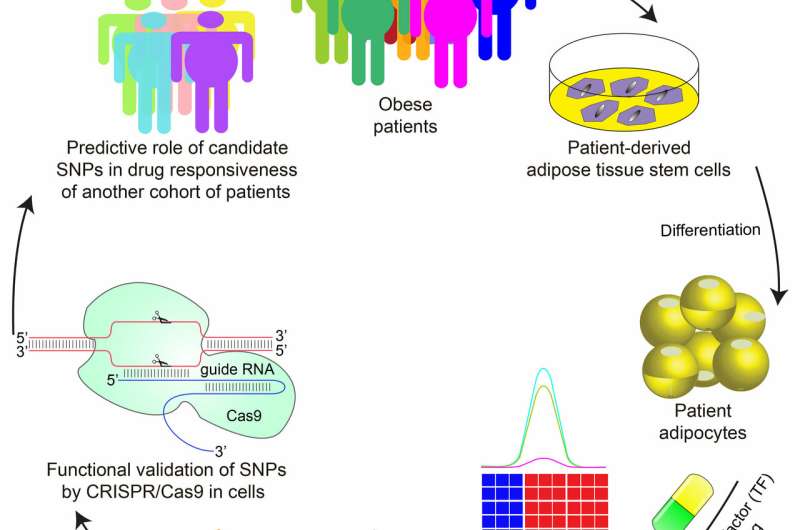
In a new study published in Cell Stem Cell, a team of researchers led by Mitchell Lazar, MD, Ph.D., director of the Institute for Diabetes, Obesity, and Metabolism in the Perelman School of Medicine at the University of Pennsylvania, have demonstrated—using fat cells derived from human stem cells—that individual genetic variation can be used to predict whether the TZD rosiglitazone will produce the unwanted side effect of increasing cholesterol levels in certain individuals.
"Obesity has reached global epidemic proportions and is a major risk factor for type 2 diabetes, so any steps we can take toward understanding how patients respond to treatment is crucial," Lazar said. "We found that genetic variation impacts how PPARγ [a receptor protein] interacts with the genome DNA in fat cells," Lazar said. "This determines an individual's responses to anti-diabetic drugs and has direct implications for developing personalized therapies for diabetes."
PPAR? is required for the maturation of fat cells and is the target of TZDs, the only class of drugs that reverse insulin resistance associated with type 2 diabetes.
By studying the genome of fat cells derived from patients treated at Penn Medicine, Lazar's team discovered a genetic variation that predicted whether rosiglitazone would increase expression of a gene called ABCA1, which regulates cholesterol levels. The variation does not appear in the protein-coding region of the ABCA1 gene, but is found in regions that code for molecules that regulate the level of ABCA1 expression. The team demonstrated the causal relationship between the genetic variation and increased ABCA1 expression by editing the variant from its inactive form to the active one using CRISPR/Cas9.
The ability of the active variant to predict whether rosiglitazone treatment will increase cholesterol was confirmed by studying 84 patients treated with the drug in Shanghai. While the overall effects of TZDs are clearly governed by many genes, the Penn study showed that individual genetic variations can predict effects of drugs on gene expression and metabolic physiology.
"Understanding the underlying mechanism driving a differential response to TZDs could inform personalized and precision approaches to treating type 2 diabetes," Lazar said. "Doctors could test for this genetic variation and choose to avoid rosiglitazone treatment of diabetic patients whose genes predict that the drug would have a greater chance of increasing their serum cholesterol levels."
These principles can be applied not only to TZDs but more broadly to other classes of drugs that work at non-coding regions of the genome, including those targeting steroid hormone receptors. The hope is to be able to predict which patients will have beneficial versus detrimental responses to drugs for tailoring drug therapy for individuals.