Scientists use crabs to validate popular method to identify unknown human brain neurons

"Being crabby" might have a whole new meaning.
A crab's nervous system could help scientists learn what causes single neurons in the human brain to become "out of whack," which can contribute to the development of neurological diseases like Alzheimer's disease. Knowing exactly how a single neuron operates among the billions housed in the human brain could one day help scientists design innovative ways to prevent and treat these diseases, such as targeted therapies.
The study, conducted by researchers at the University of Missouri, Brandeis University and the University of Texas at Austin, was published in the journal Proceedings of the National Academy of Sciences.
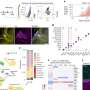
Researchers worked to validate a popular research method called RNA sequencing used to identify unknown neurons in the brain and sort them into various subtypes. Neurons are a basic element of all animal nervous systems, allowing scientists to draw comparisons in animal models like crabs when studying the human nervous system.
"There are billions of neurons in the human brain, yet we still don't know how many distinct types there are," said David Schulz, a professor of biological sciences in the College of Arts and Science. "We are finally at a technological point where we can ask the incredibly complex question—what are the brain's building blocks?"

Schulz believes the answer to that question will drive everything we know about diseases in the brain for the next 50 to 100 years. However, in order to answer that question he said we must first know how neurons are different from one another, and how healthy neurons differ from diseased ones.
Using a crab's nervous system as a model, the researchers compared and validated the results of previous human RNA sequencing methods. Since crabs have already identifiable subtypes of neurons, the researchers knew what they were looking for, so they were able to work backward from the published results and use the RNA sequencing method to validate those findings.
Schulz said he was both surprised and reassured by what they found.
"If you don't know what you are looking for in the complex human brain, then early efforts using RNA sequencing are going to need some refinement before we can answer this fundamental question," Schulz said. "This study is one of those refinements. Until we can understand each component, we can't expect to take the brain apart and put it back together again in order to figure out how it works."
The study, "Molecular profiling of single neurons of known identity in two ganglia from the crab Cancer borealis," was published in the Proceedings of the National Academy of Sciences.
More information: Adam J. Northcutt et al, Molecular profiling of single neurons of known identity in two ganglia from the crab Cancer borealis, Proceedings of the National Academy of Sciences (2019). DOI: 10.1073/pnas.1911413116